PROJECTS
Reimagining Interpretability
maxATAC
HEp-2 Benchmarking
NeuroGleam
Gait Estimation
Deepimmuno
Bearing Classifier
Reimagining Interpretability in Segmentation: A Multi-Scale Coherence Framework
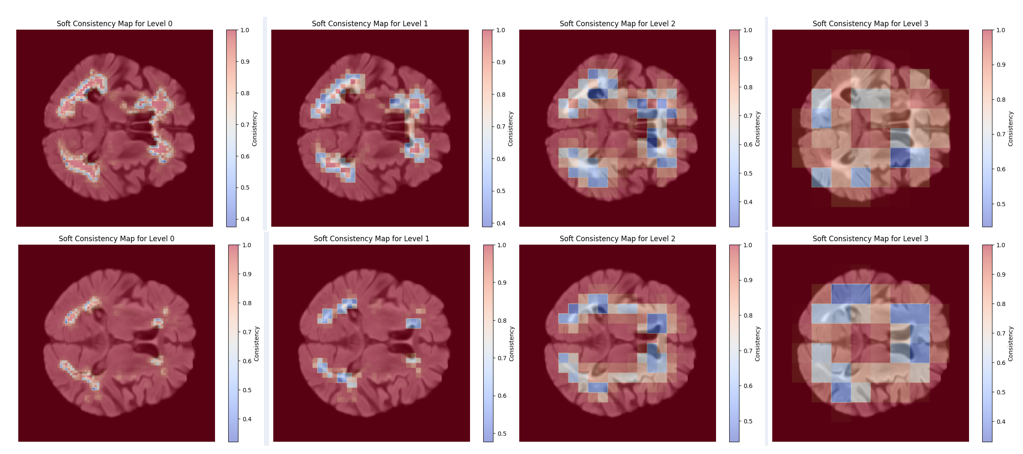
Ever wonder how your AI model really “sees” an image beneath the final segmentation map? In this project, we introduce a novel multi-scale coherence framework that illuminates the hidden decision-making process inside deep learning models—particularly where they might be uncertain or unreliable.
Using an XOR-based Deep Supervision (XDS) technique, our approach peels back the layers of segmentation from coarse to fine resolution. We then measure cross-scale consistency and visualize exactly where the model’s predictions align or deviate. Think of it as giving your segmentation model an on-the-fly “lie detector,” revealing hotspots of confusion at boundary regions, detecting out-of-distribution inputs, and offering new metrics (like Transition Dice Score and Outlier Consistency Score) for trustworthiness.
Our method is computationally and architecturally lightweight (no major architectural overhauls needed) and domain-agnostic—we’ve demonstrated it on brain MRI and cell microscopy data, but it can be applied to virtually any segmentation task that demands reliability and interpretability.
Key Highlights
- XDS Phased Training:A curriculum-inspired pipeline that refines segmentation from coarse to fine scales.
- Heatmaps of Uncertainty: Multi-scale coherence maps that show exactly where and why the model struggles (e.g., tricky organ/lesion boundaries in MRI)
- Practical OOD Detection: Identify out-of-distribution scans by tracking consistency across resolutions—critical for robust clinical or industrial AI deployment.
Curious to see how it all works? |
Paper |
Code |
maxATAC: Genome-Scale transcription factor binding prediction from ATAC-Seq with deep neural networks

While genes are often considered the primary drivers of health and disease, many crucial mutations reside in noncoding regions—regulatory DNA sequences that determine when and where genes are activated. These regions are interpreted by transcription factors (TFs), specialized proteins that read DNA sequence to orchestrate gene expression in a highly cell-specific manner. However, systematically mapping TF binding sites across diverse cell types is a daunting challenge. maxATAC is a deep learning framework that harnesses ATAC-seq data to predict TF binding sites with remarkable accuracy. By integrating chromatin accessibility with deep neural networks, maxATAC unveils how genetic variants and cellular contexts influence gene expression and impact disease risk and progression.
We built maxATAC, the most comprehensive suite of deep neural network models for transcription factor (TF) binding prediction—spanning 127 human TFs and counting! Unlike conventional motif scanning, which relies on static sequence patterns, maxATAC leverages real-world ATAC-seq datasets to train models that recognize cell-type-specific chromatin accessibility. Our approach integrates multi-cell-type training, deep convolutional architectures, and advanced data augmentation strategies to generalize TF binding predictions across diverse cellular contexts. Whether applied to bulk or single-cell ATAC-seq, maxATAC infers TF occupancy with unprecedented precision, enabling researchers to explore gene regulatory mechanisms in both health and disease.
Key Highlights
- Wide Coverage:Genome-scale coverage for 127 TFs, each with a specialized deep CNN model.
- Cross-cell-type robustness: MPredicts TF occupancy even in unseen primary cells or single-cell data.
- Beyond motifs: Learns context-dependent features missed by basic motif scanning, yielding more accurate and insightful binding maps.
With maxATAC, uncovering where TFs bind in living cells becomes more precise, scalable, and easier than ever. If you’re digging into transcriptional regulation, complex disease genetics, or single-cell studies, consider giving maxATAC a spin!
Learn More: |
Paper |
Code |
Benchmarking HEp-2 Cell Segmentation Methods in Indirect Immunofluorescence Images - Standard Models to Deep Learning
Indirect Immunofluorescence (IIF) is a gold-standard technique for detecting autoimmune diseases, relying on HEp-2 (Human Epithileal) cell imaging to identify autoantibodies linked to conditions such as lupus, rheumatoid arthritis, and multiple sclerosis. However, manual assessment of these fluorescence patterns is time-consuming and subject to variability, necessitating automated solutions for accurate and reproducible analysis.
In this study, we conducted an extensive benchmarking of deep learning-based segmentation models for HEp-2 cell analysis, comparing 17 CNN-based architectures and 2 transformer-based models under diverse training conditions. Our experiments, leveraging the I3A dataset and rigorous statistical validation, establish baseline performance and highlight key challenges such as class imbalance and annotation quality. To address these issues, we evaluated advanced pretraining strategies, data augmentation techniques, and generative adversarial networks (GANs) for improving segmentation performance, particularly for underrepresented staining patterns.
Our insights not only highlight the current state of HEp-2 cell segmentation but also provide a roadmap for future research, guiding the development of more sophisticated and robust deep learning models capable of overcoming the unique challenges posed by HEp-2 cell analysis.
Learn More: |
Paper |
Code |
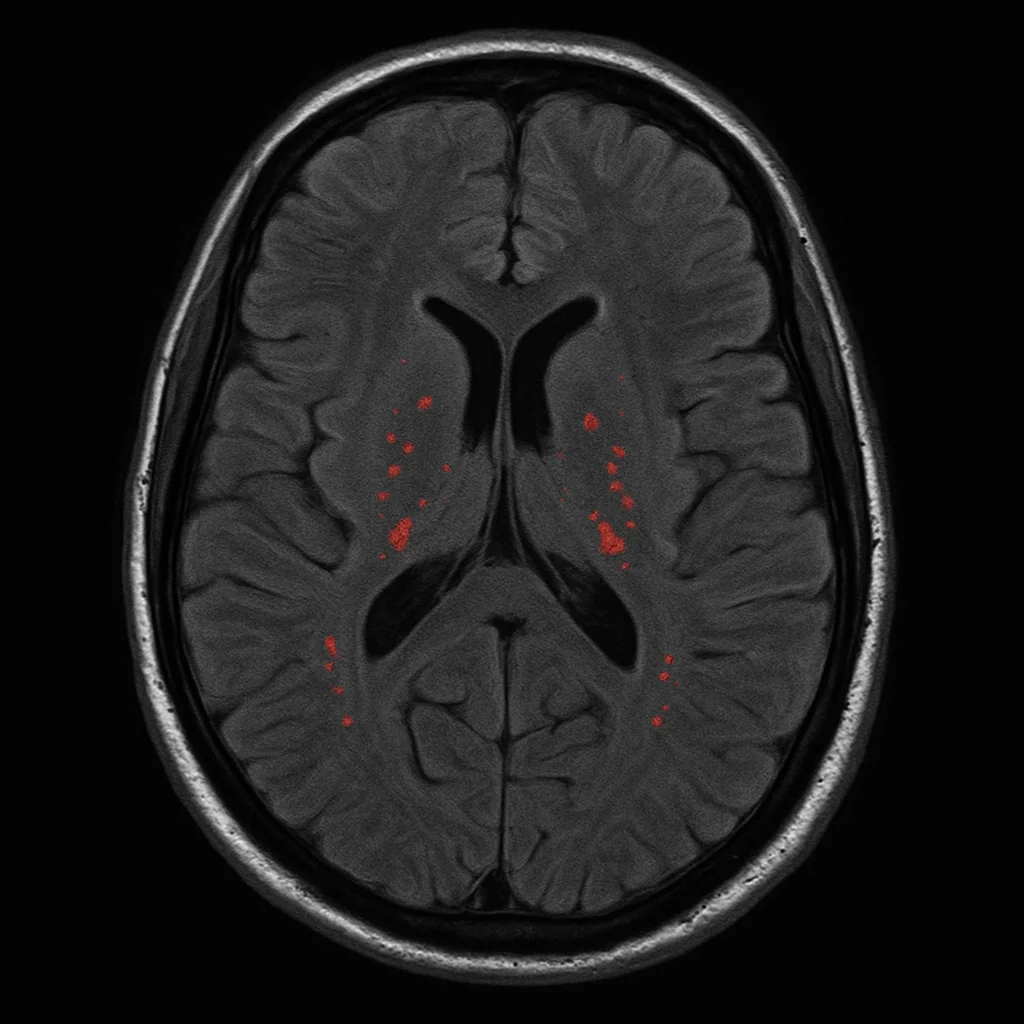
NeuroGleam: Illuminating Small Vessel Disease Detection through Deep Learning based Segmentation of Brain MRI White Matter Hyperintensities
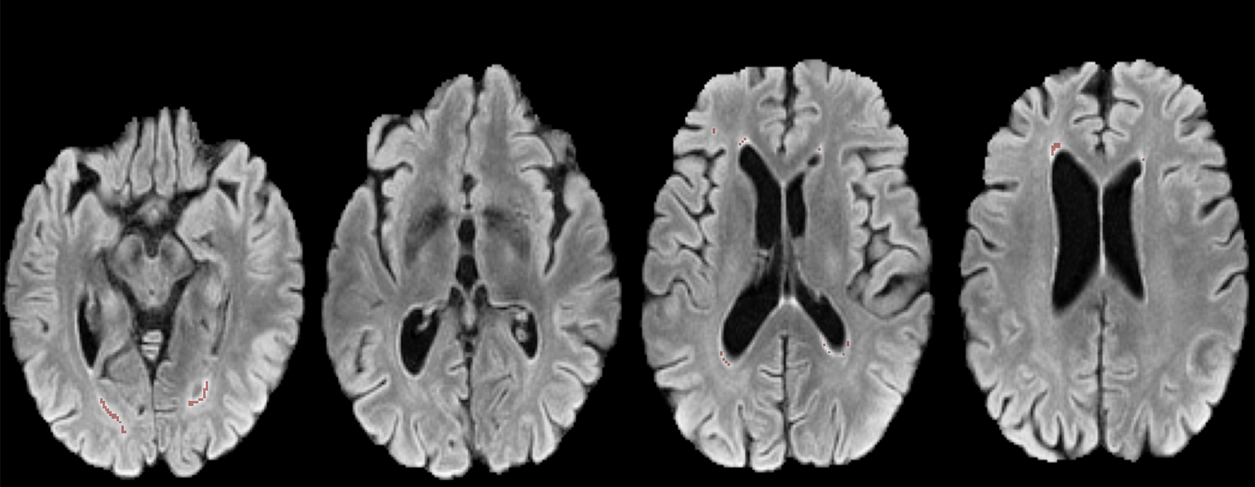
White matter hyperintensities (WMHs) are a key neuroimaging biomarker for cerebral small vessel disease (SVD) and age-related brain changes. Yet, their accurate detection in clinical MRI scans—especially using low-resolution T2-FLAIR images—remains a major challenge. NeuroGleam tackles this by developing and benchmarking deep learning models that segment WMHs with precision, scalability, and clinical applicability.
We systematically evaluated a wide range of architectures—including U-Net variants and HRNet-based models—across two diverse datasets: the research-grade MICCAI WMH Challenge dataset and the real-world APRlSE clinical dataset. Our pipeline included patch-based processing, advanced loss functions like Focal Tversky Loss, and Bayesian hyperparameter optimization to improve both accuracy and generalizability. Despite strong in-domain performance (Dice scores up to 0.682), our findings underscore the significant performance drop under domain shifts—highlighting the urgent need for domain-adaptive models for clinical use.
NeuroGleam lays the foundation for robust, real-world WMH segmentation tools that could enhance diagnostic workflows for SVD, stroke risk, and cognitive decline in everyday radiology settings.
Learn More: |
Paper |
Code |
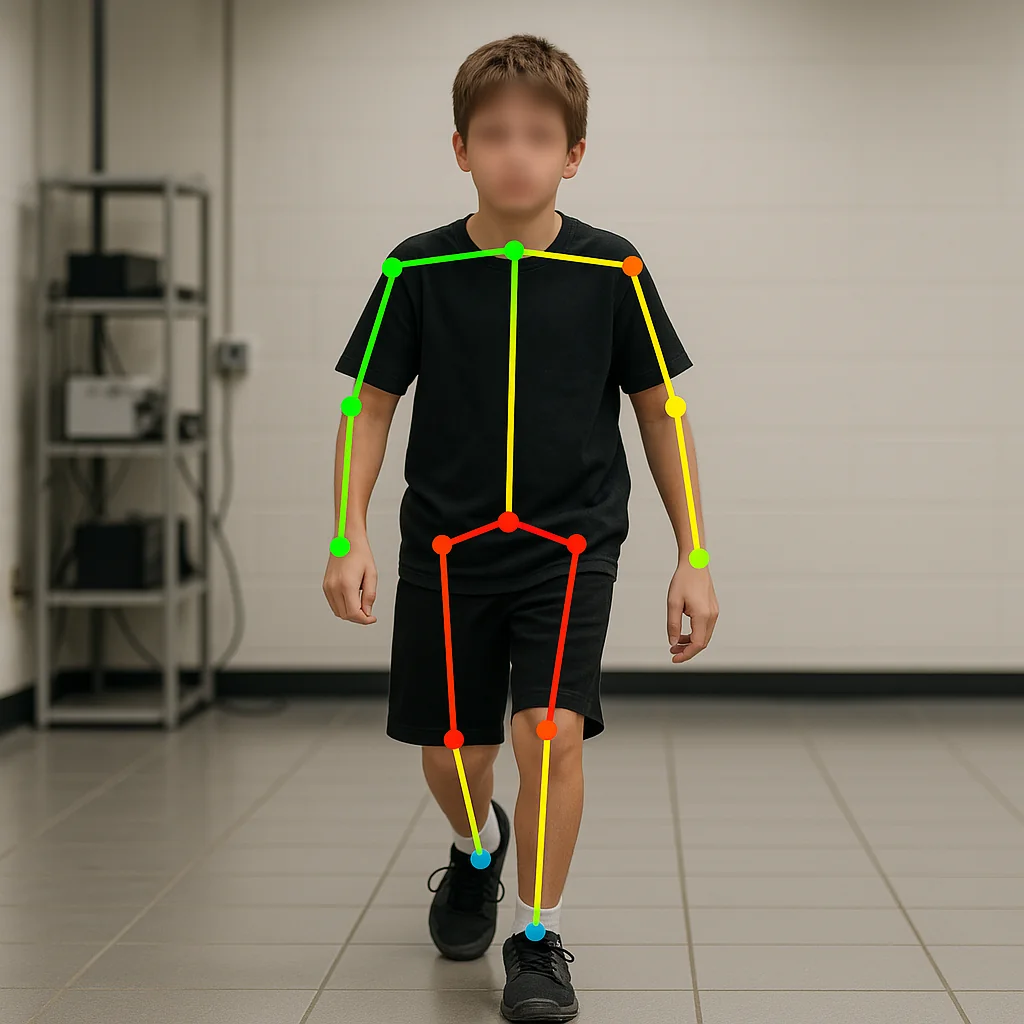
AI-driven Gait Parameters Estimation from Videos for Cerebral Palsy Patients
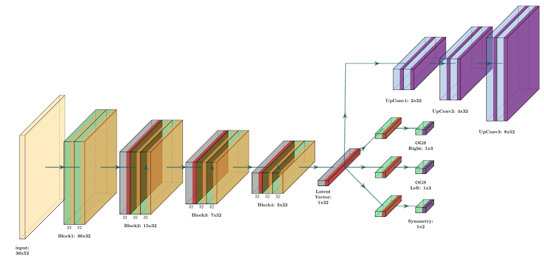
Gait analysis is critical for managing Cerebral Palsy (CP)—a condition marked by motor dysfunction affecting posture and movement. Traditionally performed in costly, sensor-equipped motion labs, clinical gait assessments are often out of reach for many patients due to logistical and financial constraints.
This project reimagines gait analysis using AI. By processing low-resolution videos from regular smartphone cameras, we estimate gait impairment using Observational Gait Score (OGS) — without the need for wearable sensors or specialized environments. Our pipeline combines pose estimation (via BlazePose), semantic feature extraction, and a deep learning model trained to predict limb-specific OGS and symmetry scores from real-world pediatric videos.
Despite challenges like background distractors and non-cooperative subjects, our models show promising results and demonstrate the clinical potential of AI-powered, low-cost gait monitoring for enhancing care accessibility and continuity in CP patients.
Learn More: |
Paper |
Code |
DeepImmuno: deep learning-empowered prediction and generation of immunogenic peptides for T-cell immunity
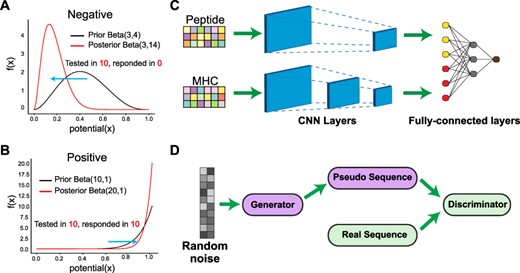
The immune system’s ability to detect threats hinges on T cells recognizing short peptides—called epitopes—presented by MHC molecules on the surface of infected or abnormal cells. Accurately predicting which peptides can trigger T-cell responses is central to designing vaccines, cancer immunotherapies, and diagnostic tools. Identifying immunogenic peptides is experimentally costly and biologically complex.
DeepImmuno is a deep learning framework that not only predicts immunogenicity with high accuracy, but also generates new peptide candidates using GAN (Generative Adversarial Network). Built on a robust beta-binomial statistical foundation, DeepImmuno learns from thousands of experimentally validated peptide–MHC examples to forecast which epitopes will activate T cells, and why. Its generative module, DeepImmuno-GAN, creates realistic peptides that mirror known antigens in key biochemical and structural features.
Tested on SARS-CoV-2 and tumor neoantigen datasets, DeepImmuno consistently outperforms state-of-the-art models and provides insights into critical TCR-contacting residues. With both a web interface and open-source code, it's a powerful platform for immune research and translational medicine.
Learn More: |
Paper |
Code |
Web Interface |
Building Classifier for Streaming Data
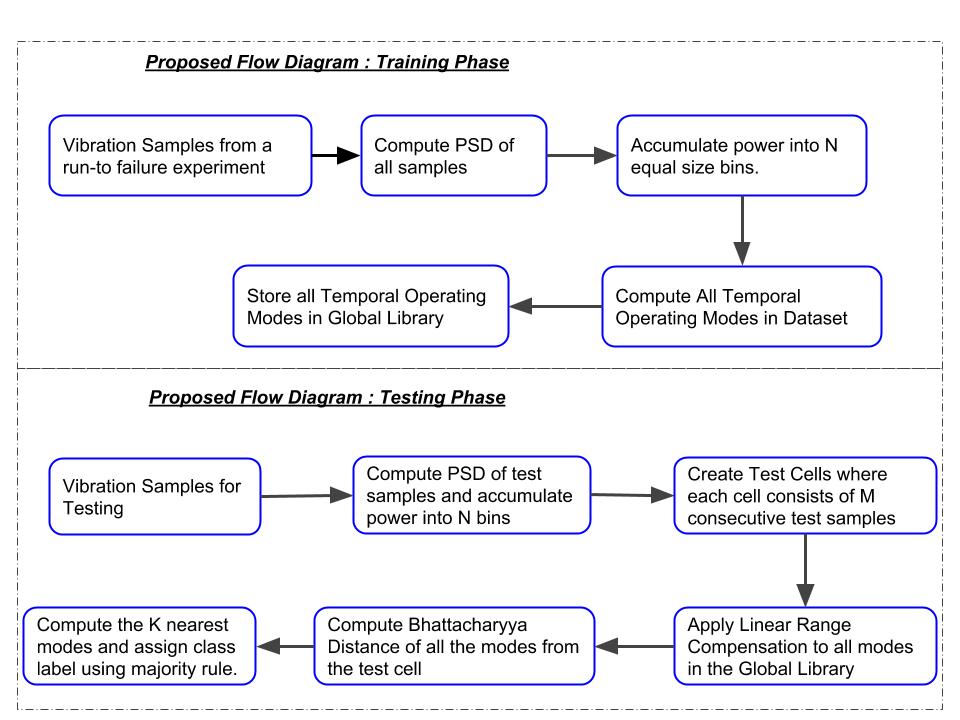
Bearings are the heartbeat of rotating machinery in manufacturing systems, and their failure can lead to costly downtimes and equipment damage. Traditional preventive maintenance methods often fall short in capturing the nuanced degradation behaviors of bearings in real-world settings. In this study, we present a novel classification framework that transforms bearing failure prediction into a binary classification problem — identifying whether a bearing failure is imminent within three weeks.
Utilizing a streaming data paradigm, the proposed method leverages vibration signal processing, Welch’s power spectral density (PSD) estimation, and Bhattacharyya distance-based similarity measures to identify temporal operating modes (TOMs). These TOMs act as behavioral landmarks, capturing subtle shifts in bearing dynamics across time. Our classifier adapts to new data via a linear range compensation mechanism, enabling accurate cross-dataset predictions even under varying operational conditions.
Experimental results demonstrate high accuracy and recall compared to traditional SVM baselines, particularly when tested across diverse degradation scenarios. This work lays the groundwork for scalable, adaptive health monitoring systems capable of real-time decision-making in industrial IoT and smart manufacturing environments.
Learn More: |
Paper |
Code |